
creating spline relativity plots
Jared Fowler
spline_pretty_relativities.RmdTo highlight some more of prettyglm’s functionality we
will now build a logistic regression model with a spline.
Pre-processing
A critical step for this package to work is to set all categorical predictors as factors. Before we fit a spline we will fit the age variable as a categorical variable to understand the trend.
library(dplyr)
library(prettyglm)
data('titanic')
# Easy way to convert multiple columns to a factor.
columns_to_factor <- c('Pclass',
'Sex',
'Cabin',
'Embarked',
'Cabintype')
meanage <- base::mean(titanic$Age, na.rm=T)
titanic <- titanic %>%
dplyr::mutate_at(columns_to_factor, list(~factor(.))) %>%
dplyr::mutate(Age = base::ifelse(is.na(Age)==T,meanage,Age)) %>%
dplyr::mutate(Age_Cat = prettyglm::cut3(Age, levels.mean = TRUE, g =10)) %>%
dplyr::mutate(Fare_Cat = prettyglm::cut3(Fare, levels.mean = TRUE, g =10))
# Build a basic glm
survival_model <- stats::glm(Survived ~ Pclass +
Sex +
Sex:Fare_Cat +
Age_Cat +
Embarked +
SibSp +
Parch,
data = titanic,
family = binomial(link = 'logit'))Understanding the trend
By setting plot_factor_as_numeric equal to TRUE in
pretty_relativities() we can plot understand the bucketed
categorical predictors trend. Setting
plot_factor_as_numeric equal to TRUE also works for
interactions like Sex:Fare.
We will fit a knot point at 18 and 35.
pretty_relativities(feature_to_plot= 'Age_Cat',
model_object = survival_model,
relativity_label = 'Liklihood of Survival',
plot_factor_as_numeric = T
)We will fit a knot point at 55.
pretty_relativities(feature_to_plot= 'Sex:Fare_Cat',
model_object = survival_model,
relativity_label = 'Liklihood of Survival',
plot_factor_as_numeric = T,
iteractionplottype = 'colour',
facetorcolourby = 'Sex')Creating the spline
prettyglm includes some useful functions to assist in
building splines. splineit() takes the minimum and maximum
points of a spline to create the splined column. An example workflow
using dplyr is shown below.
titanic <- titanic %>%
dplyr::mutate(Age_0_18 = prettyglm::splineit(Age,0,18),
Age_18_35 = prettyglm::splineit(Age,18,35),
Age_35_120 = prettyglm::splineit(Age,35,120)) %>%
dplyr::mutate(Fare_0_55 = prettyglm::splineit(Fare,0,55),
Fare_55_600 = prettyglm::splineit(Fare,55,600))
survival_model4 <- stats::glm(Survived ~ Pclass +
Sex:Fare_0_55 +
Sex:Fare_55_600 +
Age_0_18 +
Age_18_35 +
Age_35_120 +
Embarked +
SibSp +
Parch,
data = titanic,
family = binomial(link = 'logit'))Visualising the spline
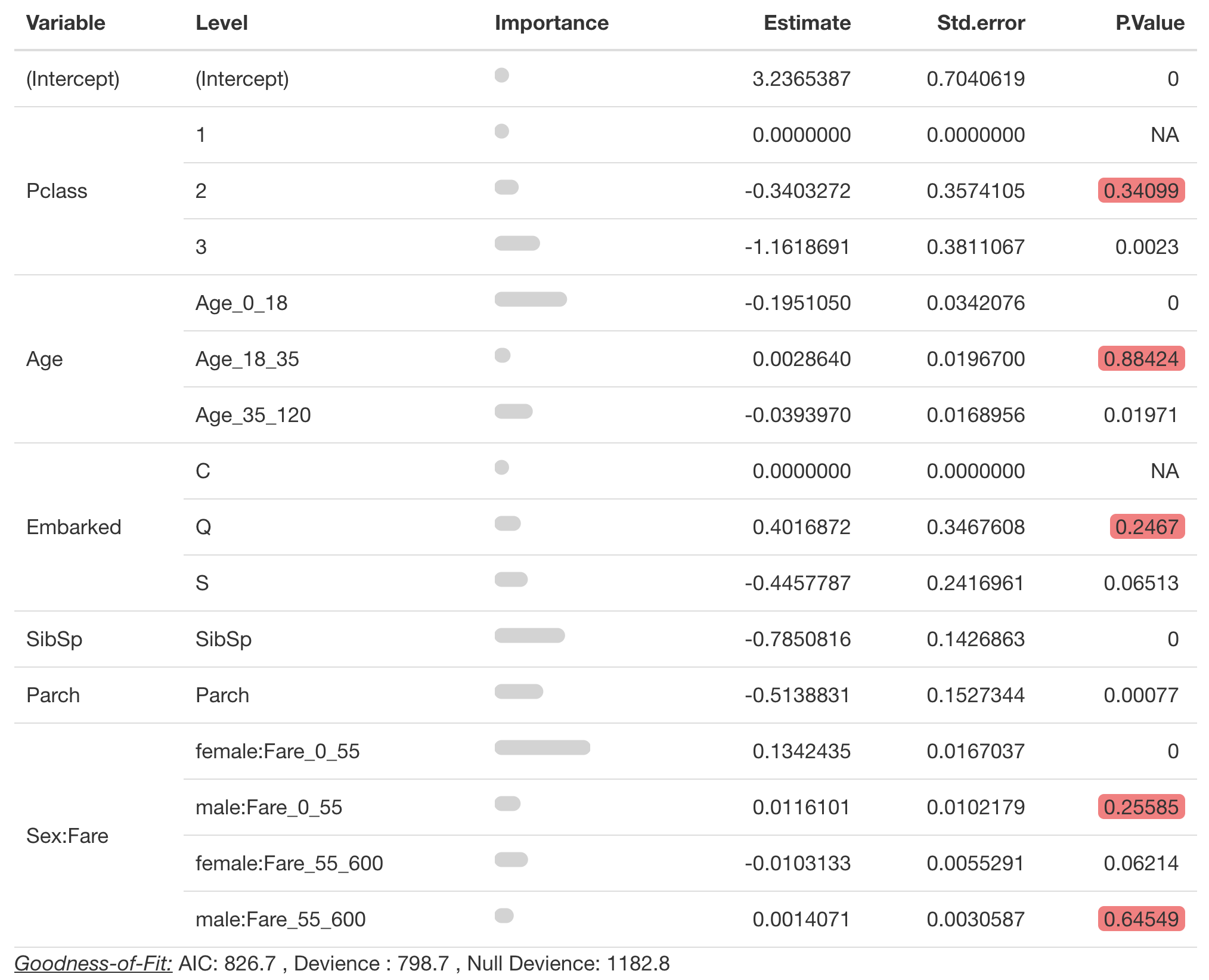
Creating a table of model coefficients
For interactions variables are grouped on the left pane under
Variables. It is important to provide the correct
spline_seperator
pretty_coefficients(survival_model4,
significance_level = 0.1,
spline_seperator = '_')
Create plots of fitted coefficients using pretty_relativities
Single Variable Splines
You also need to provide a spline_seperator input in
pretty_relativities().
pretty_relativities(feature_to_plot= 'Age',
model_object = survival_model4,
relativity_label = 'Liklihood of Survival',
spline_seperator = '_'
)Interacted Splines
By default pretty_relativities() will colour by the
factor variable.
pretty_relativities(feature_to_plot= 'Sex:Fare',
model_object = survival_model4,
relativity_label = 'Liklihood of Survival',
spline_seperator = '_',
upper_percentile_to_cut = 0.03
)If you prefer to facet by the factor variable, change
iteractionplottype to “facet”
pretty_relativities(feature_to_plot= 'Sex:Fare',
model_object = survival_model4,
relativity_label = 'Liklihood of Survival',
spline_seperator = '_',
upper_percentile_to_cut = 0.03,
iteractionplottype = 'facet'
)