Creating a coefficient table
Jared Fowler
creating_pretty_coefficients.RmdThe function pretty_coefficients() allows you to create
a pretty table of model coefficients. The table created will
automatically include categorical variable base levels, and highlight
insignificant p-values.
Pre-processing
A critical step for this package to work is to set all categorical predictors as factors.
library(dplyr)
library(prettyglm)
data('titanic')
# Easy way to convert multiple columns to a factor.
columns_to_factor <- c('Pclass',
'Sex',
'Cabin',
'Embarked',
'Cabintype')
meanage <- base::mean(titanic$Age, na.rm=T)
titanic <- titanic %>%
dplyr::mutate_at(columns_to_factor, list(~factor(.))) %>%
dplyr::mutate(Age =base::ifelse(is.na(Age)==T,meanage,Age))
# Build a basic glm
survival_model <- stats::glm(Survived ~ Pclass +
Sex +
Fare +
Age +
Embarked +
SibSp +
Parch,
data = titanic,
family = binomial(link = 'logit'))Table of model coefficients
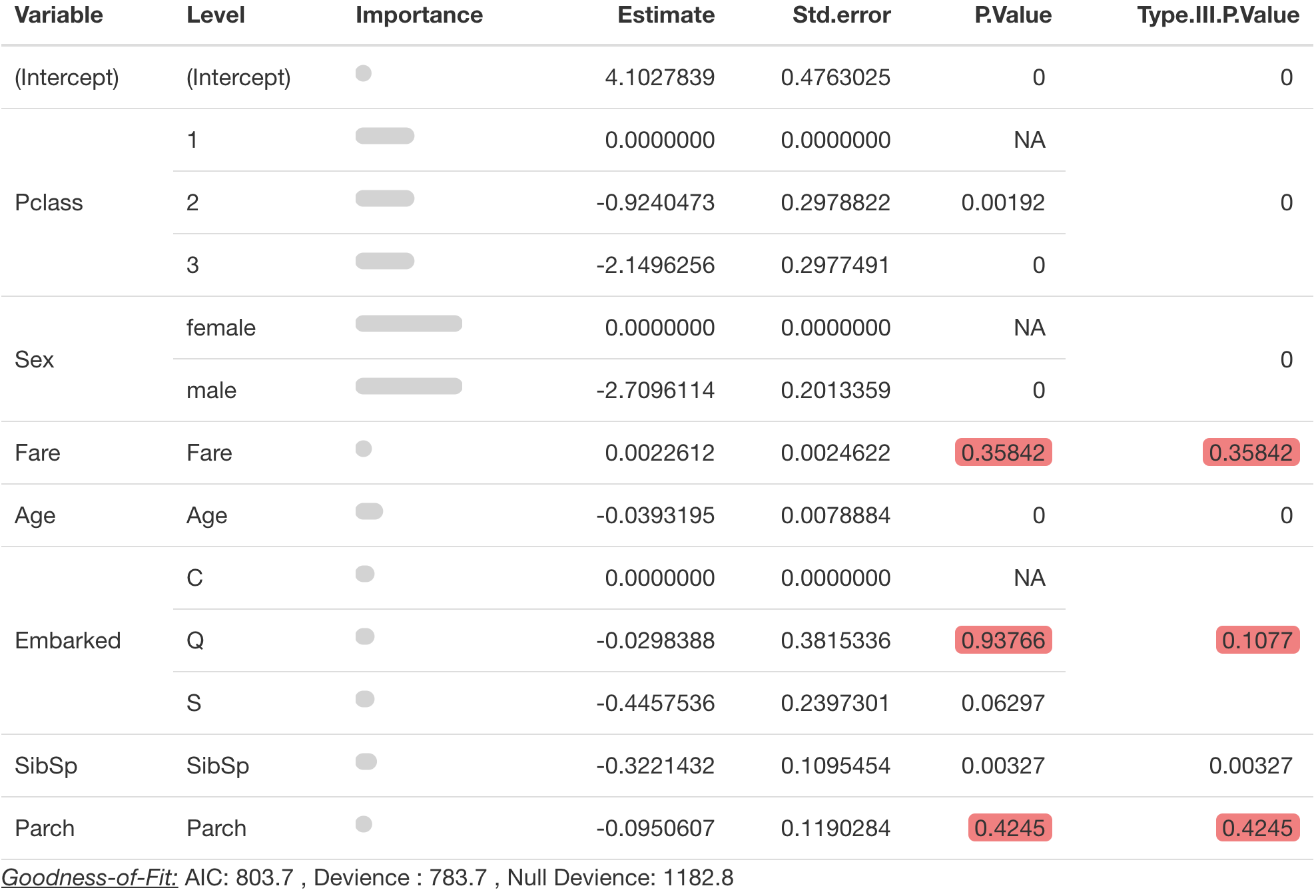
The simplest way to call this function is just with the model object.
pretty_coefficients(model_object = survival_model)
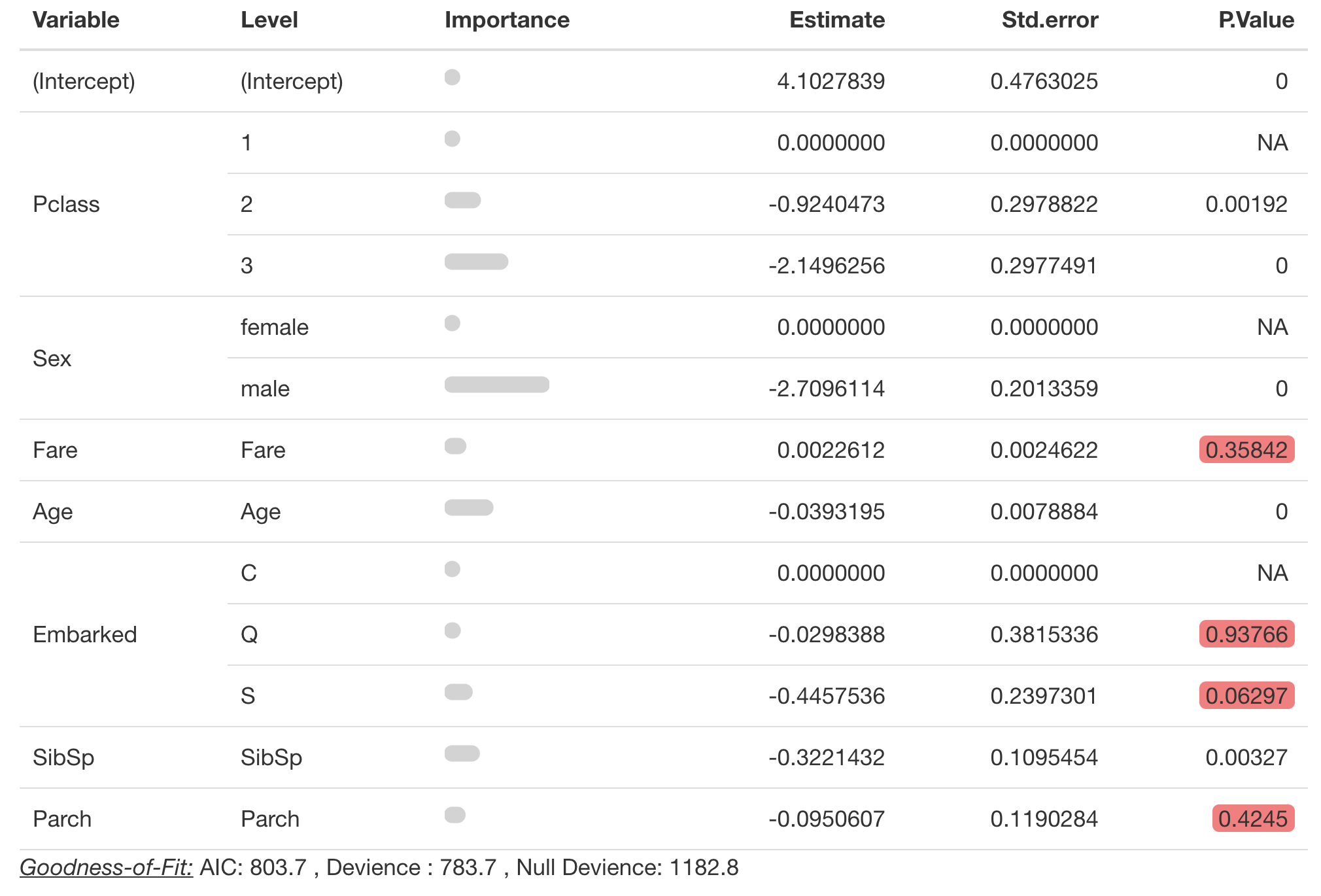
You can also complete a type III test on the coefficients by
specifying a type_iii argument. Warning Wald
type III tests will fail if there are aliased coefficients in the
model.
You can change the significance level highlighted in the table with
significance_level.
pretty_coefficients(survival_model,
type_iii = 'Wald',
significance_level = 0.1)
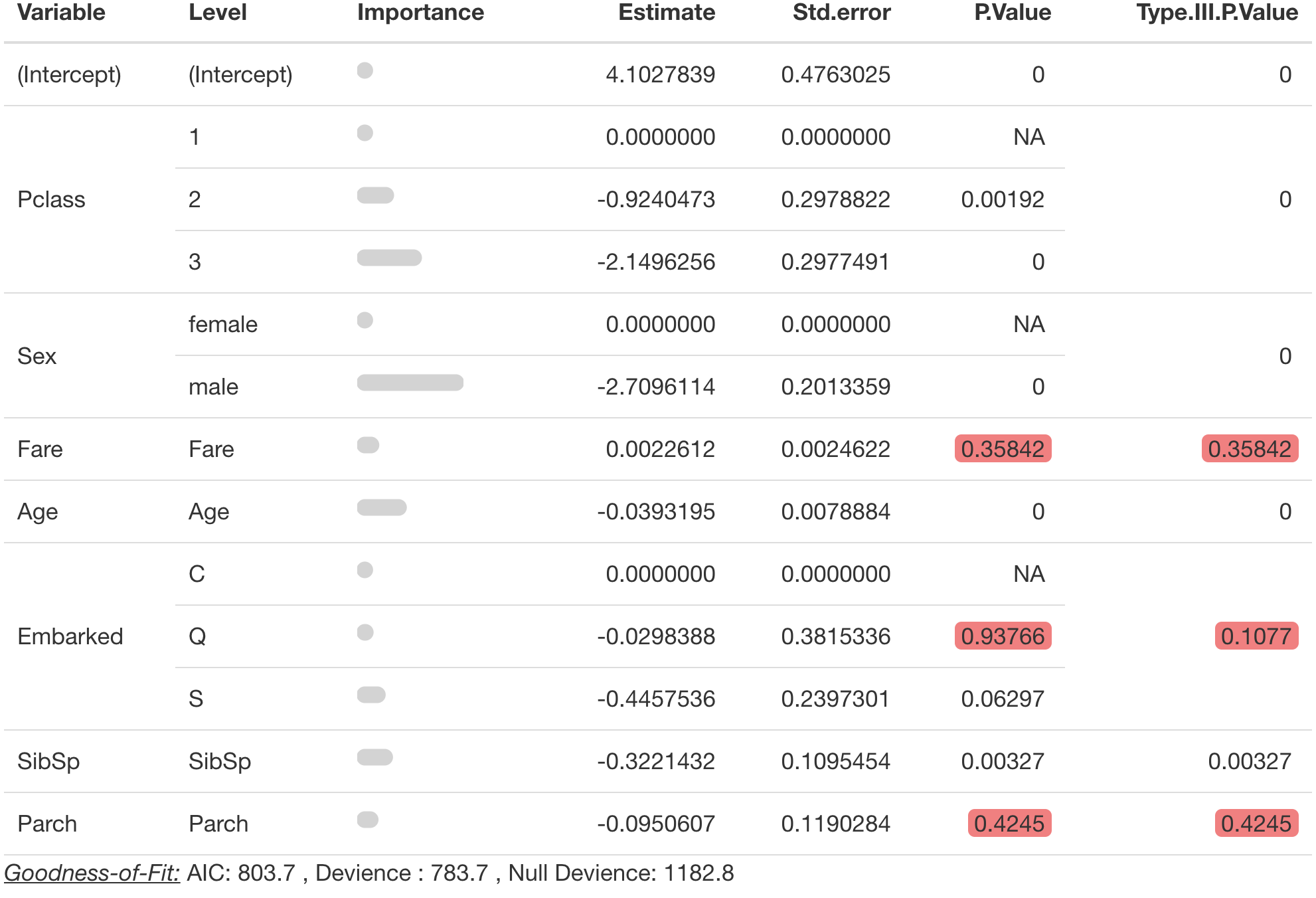
By default pretty_coefficients shows “model” variable
importance. But vimethod also accepts “permute” and “firm”
methods from . Additional parameters for these methods should also be
passed into pretty_coefficients().
pretty_coefficients(model_object = survival_model,
type_iii = 'Wald',
significance_level = 0.1,
vimethod = 'permute',
target = 'Survived',
metric = 'auc',
pred_wrapper = predict.glm,
reference_class = 0)